- Home
- About
- OSD
- MyOSD
- Partners
- Work Packages
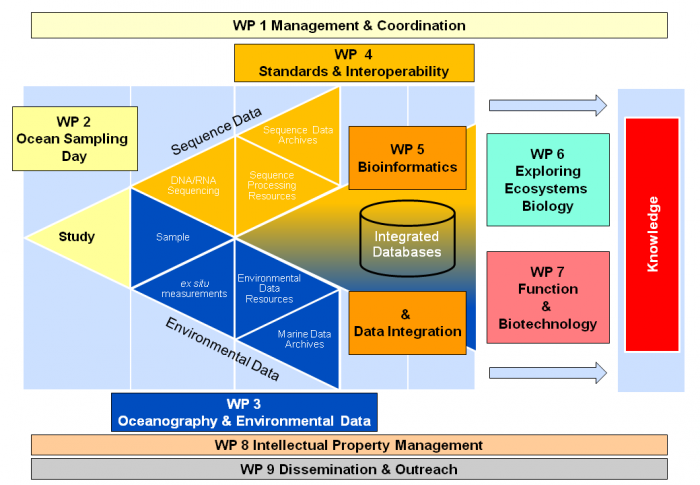
- WP 1Management & Coordination
- WP 2 (OSD)Ocean Sampling Day
- WP 3Oceanography & Environmental Data
- WP 4Standards and Interoperability
- WP 5Bioinformatics & Data Integration
- WP 6Exploring Ecosystems Biology
- WP 7Function and Biotechnology
- WP 8Intellectual Property (IP) Management for Marine Bioprospecting
- WP 9Dissemination & Outreach
- Public DeliverablesAll Micro B3's public deliverables
- Meetings
- Workshops
- Third Micro B3 Industry Expert Workshop
- Micro B3 Industry Expert Workshop
- Micro B3/OSD Analysis Workshop
- Micro B3 Stakeholder Workshop
- Micro B3 Summer School in Crete 2014
- Marine Metagenomics Bioinformatics
- Micro B3 Industry expert workshop
- EU-US Training 2013
- Micro B3 Statistics Training 2013
- MG4U Bioinformatics Training 2013
- Bioinformatics Training 2012
- EU-US Training 2012
Background
What is Micro B3?
 Micro B3 stands for “Microbial Biodiversity, Bioinformatics and Biotechnology“ which reflects the three main pillars the project addresses. The central concept of this collaborative project (large-scale integrating project) is to team up an interdisciplinary and intersectoral set of world class European researchers to overcome current obstacles in marine biodiversity research and blue biotechnology. Micro B3 fully addresses the Ocean of Tomorrow Call Topic OCEAN.2011-2: “Marine microbial diversity – new insights into marine ecosystems functioning and its biotechnological potential”.
Micro B3 stands for “Microbial Biodiversity, Bioinformatics and Biotechnology“ which reflects the three main pillars the project addresses. The central concept of this collaborative project (large-scale integrating project) is to team up an interdisciplinary and intersectoral set of world class European researchers to overcome current obstacles in marine biodiversity research and blue biotechnology. Micro B3 fully addresses the Ocean of Tomorrow Call Topic OCEAN.2011-2: “Marine microbial diversity – new insights into marine ecosystems functioning and its biotechnological potential”.
Why does Europe need Micro B3?
The Planet Earth is a blue planet – with 70 % of its surface covered by oceans and 40 % of the World’s population living within 50 km of a coastline, our heritage, economy and wellbeing are inextricably linked with the marine environment. Marine microbes are the ‘gatekeepers’ for the Earth System with an estimated contribution to global primary productivity between 50% to 90%. Recent investigations have shown that beside the classic carbon and energy flow through photosynthetic Eukarya to herbivores and on to higher trophic levels, a rather effective microbial food web exists. Autotrophic microorganisms fix carbon and - in case of Cyanobacteria - release oxygen; heterotrophic microorganisms use energy stored in the non-living, detrital carbon pool, recycling nitrogen, sulphur and phosphorus and other elements. Additionally, there is ample evidence that marine heterotrophic bacteria can use light as an additional energy source to survive under conditions of nutrient starvation and that viruses are potential mediators of the microbial life.
Although about half of the annual primary production of the planet occurs in the ocean and bacterial metabolism is involved in the chemical transformation of most elements, very little is known of marine microbial diversity. The vast majority (90-99%) of these organisms can not be retrieved as axenic cultures under standard laboratory conditions and so are not amenable to study by the classical methods that have proved so successful throughout the 20th century. It was only with the development of a molecular toolbox to sequence DNA from the natural environment that information about the exceptional amount of biodiversity and function in the oceans began to accumulate.
In recent years marine ecosystems research has benefitted greatly from the use of genomic (sequencing of axenic cultures) and metagenomic (sequencing of community DNA) approaches. In fact DNA-based sequencing, originally developed in the biomedical field, has been quick to be incorporated in the marine sciences. In particular, metagenomics has resulted in an explosion of information on marine microbes. For example, the first part of the Global Ocean Sampling (GOS), which sampled the North Atlantic, Caribbean and a small part of the Pacific Ocean, added DNA sequence information that was equivalent to 50% of all protein-encoding sequences that had previously been deposited in the public repositories of the International Nucleotide Sequence Database Collaboration (INSDC). GOS confirmed that marine microbes are diverse, revealing how little is known about the genetic information of natural assemblages.
For the first time in biology the new high throughput achieved by next generation sequencing technologies (NGS) promise to overcome this gap in knowledge by providing information on the genetic diversity and potential function of the organisms on an unprecedented scale. Sequencing a genome or metagenomes has become a simple and cheap component of any study. New third generation ultra-high-throughput sequencing technologies currently entering the market, promise even cheaper per base prices enticing researchers further to scale up their sequencing efforts. However, the processing and analysis of the data mostly outcompetes the bioinformatic capacities of many researcher groups and institutes in the marine field. In fact the new sequencing machines are described to bring what has been termed disruptive technology for scientists all over Europe (meaning that the rate at which it improves exceeds the rate users can adapt to the new performance). One of the reasons is that only in the US cyber-infrastructures have been developed to be able to cope with large-scale data analysis. The CAMERA system, in particular, was designed to meet the challenge of studying marine life and ecosystems for examining the genomic complexities of natural communities of microorganisms as they have evolved in their local environments. Further multi-purpose analysis pipelines in the US are IMG/M and MG-RAST. Although very useful also for European researchers, they do not provide the capacities, flexibility and a higher level of data integration framework that is needed to complement the ecosystems expertise of marine ecologists with appropriate bioinformatic approaches, finally widening the gap between technology providers and field researchers.
This again demonstrates that technological capacities alone do not provide much information when it comes down to a better understanding of the diversity and function of microorganisms. But this is a prerequisite for inferring an organism’s ecology, as well as for discovering new catalytic mechanisms and enzymes.
Micro B3: By concept interdisciplinary, intersectoral and integrative!
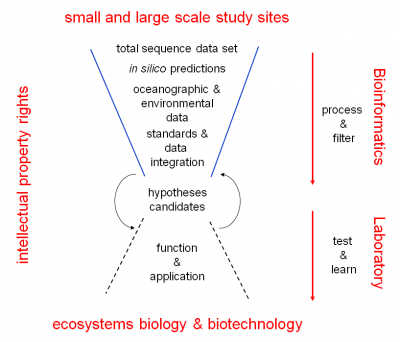 Micro B3 will address these issues by polling Europeans’ expertise for the development of new bioinformatic approaches to analyse, integrate and visualize marine molecular and environmental data jointly. From the beginning on this will be done in close collaboration with field scientists providing ecosystems expertise as well as their small and large-scale data sets. In close collaboration with Genoscope and the European Bioinformatics Institute (specifically EMBL-EBI), tailored solutions will be developed for data analysis, representation and transfer. To move forward in better understanding the marine ecosystem Micro B3 will go far beyond providing only solutions for initial data processing and preparation for storage. Micro B3 will for the first time follow a holistic approach that integrates the information on the diversity of organisms with their potential functions, reflected in their genes, and with local as well as global environmental parameters. To integrate these heterogeneous data streams, georeferencing has been shown to be extremely useful, especially for the open ocean, where any kind of diversity and genomic data can be easily linked with measured and global oceanographic parameters based on the three simple universal descriptors: location, time and depth. Important prerequisites to facilitate a high level of interoperability and exchange of data are standards for data formats, as well as are quality management to provide confidence in prediction and interpolation. Together with the Genomic Standards Consortium, an open-membership, international working body founded in 2005 to provide mechanisms that standardize the description of genomes and the exchange and integration of genomic data, and marine data initiatives and databases (EMODNet, PANGAEA, SeaDataNet, Group on Earth Observations (GEOSS), Global Monitoring for Environment and Security GMES) existing and new standards and mechanisms for seamless data transfer will be developed and deployed. The enriched and integrated datasets will be used for ecosystems modelling to reach a better understanding of the complexity of microbial communities and their role in climate change.
Micro B3 will address these issues by polling Europeans’ expertise for the development of new bioinformatic approaches to analyse, integrate and visualize marine molecular and environmental data jointly. From the beginning on this will be done in close collaboration with field scientists providing ecosystems expertise as well as their small and large-scale data sets. In close collaboration with Genoscope and the European Bioinformatics Institute (specifically EMBL-EBI), tailored solutions will be developed for data analysis, representation and transfer. To move forward in better understanding the marine ecosystem Micro B3 will go far beyond providing only solutions for initial data processing and preparation for storage. Micro B3 will for the first time follow a holistic approach that integrates the information on the diversity of organisms with their potential functions, reflected in their genes, and with local as well as global environmental parameters. To integrate these heterogeneous data streams, georeferencing has been shown to be extremely useful, especially for the open ocean, where any kind of diversity and genomic data can be easily linked with measured and global oceanographic parameters based on the three simple universal descriptors: location, time and depth. Important prerequisites to facilitate a high level of interoperability and exchange of data are standards for data formats, as well as are quality management to provide confidence in prediction and interpolation. Together with the Genomic Standards Consortium, an open-membership, international working body founded in 2005 to provide mechanisms that standardize the description of genomes and the exchange and integration of genomic data, and marine data initiatives and databases (EMODNet, PANGAEA, SeaDataNet, Group on Earth Observations (GEOSS), Global Monitoring for Environment and Security GMES) existing and new standards and mechanisms for seamless data transfer will be developed and deployed. The enriched and integrated datasets will be used for ecosystems modelling to reach a better understanding of the complexity of microbial communities and their role in climate change.
Although our current knowledge on marine microbial communities is poor, and might appear a limitation for biotechnological exploitation, we believe that Micro B3’s innovative, integrative approach offers excellent opportunities for bioprospecting for novel enzymes of industrial interest and for metabolic products. The marine environment is very diversified, ranging from cold to hot, and with hypersaline and high pressure habitats. Organisms that have evolved to occupy this wide range of ecological niches must have diverse metabolisms and will, therefore, possess novel enzyme capability. Many of the processes that these microbes can carry out are likely to be of interest to industry. The challenges in this respect are not only to search for and nail down the functions of candidate genes for biotechnological applications but to be able to assign functions to some of the up to 50% of hypothetical and conserved hypothetical genes commonly found in metagenomic applications. The tight integration with world-class experts and companies on enzyme and bio-actives discovery within Micro B3 will guarantee to match marked demands within the framework of the project. As an important step to facilitate opening up the black box of microbial diversity and function in the marine system Micro B3 will take care of the development of an appropriate legal framework to protect our marine genetic resources which in turn will allow for the sustainable use of the data and materials sampled. Marine researchers and stations, the Mediterranean Science Commission (CIESM) and the EuropaBio industry association will second the development of IP agreements for pre-competitive microbial research materials and data as well as for the exploitation of high-potential commercial applications by providing access to a large set of scientists, end-users and policy makers.
To make the developments of Micro B3 transparent and accessible for a wide range of researchers an Ocean Sampling Day will be initiated around 30 month after the project start. Organized by Micro B3, selected marine stations are asked to take samples and contribute data in a concerted action. A comprehensive registry of interesting sampling sites, compiled in the project, will be the basis for safeguarding existing data and knowledge and the organisation of this endeavour. The Ocean Sampling Day will also enforce the timely deployment of standards, best-practice solutions and protocols developed within Micro B3 as well as provide the perfect use case to test the bioinformatic developments and legal framework by model contracts for access and benefit sharing. The data gained will contribute to establish a standardized baseline for marine biodiversity and the functions they provide on the molecular level. This is an important step towards understanding the impact of marine microorganisms on global change and the effect of global change on microbial diversity. It will help in developing ecosystems models that include the microbial ‘factor’ finally leading to predictions on the capacity of marine ecosystems to respond to natural and anthropogenic changes.
The European background of Micro B3
Micro B3 has its roots in the 6 FP NEST (New Emerging Science and Technology) project MetaFunctions coordinated by MPIMM from 2005 to 2008. Already in this project the objective was to use data integration and make use of the emerging molecular datasets in the marine system to find new processes and enzymes. Although from the technological point very successful at this time, the limited amount of quality controlled and standardized sequence data and particularly contextual data (environmental parameters) led to a limited amount of additional knowledge on the marine ecosystem. It was only with the implementation of the Networks of Excellence (NoEs), Marine Biodiversity and Ecosystems Functioning (MarBef) and Marine Genomics Europe (MGE) that a larger community of marine researchers could be involved to further develop the idea of large-scale data integration in the marine system raising the need for specific bioinformatic capacities tailored for the marine community. Participation in the FP7 projects EuroFleets, EuroMarine and MAMBA (Marine Metagenomics for new Biotechnological Applications) finally paved the road to substantially team up marine researchers and technology developers with oceanography and blue biotechnology. The concept of Micro B3 includes strong links to the emerging ESFRI infrastructure projects ELIXIR, EMBRC (European Marine Biological Resource Centers), LifeWatch and EMODNet, because we believe that our developments will go far beyond the project itself and add value to these growing infrastructures for bioinformatic and marine research. Our interdisciplinary group of advisory board members will help us in this respect. In addition Micro B3 will address pressing IPR issues which have not been addressed in a comprehensive way in any other marine project before. With CIESM and a set of small to large-scale sampling initiatives being part of Micro B3 plus the expertise of EuropaBio and several biotech companies, we expect to have the right mixture to develop an appropriate legal framework and model contracts that are acceptable by researchers and companies to guarantee access to samples and data as well as proper benefit sharing. Finally, the concept of Micro B3 addresses recent recommendations in the field of blue biotechnology (Bremen meeting on Marine Biotechnology Research, 2007 and “A European strategy for marine and maritime research”, 2008) as well as recommendations of the EC-US Task Force on Biotechnology Research (Joint CIESM/EC-US meeting, Monaco 2008) to “strengthen Europe’s bioinformatics research capability”.
Objectives of Micro B3
The major objectives of the Micro B3 project are:
1. To create a new inter- and multidisciplinary culture in marine sciences comprising:
a. State-of-the-art facilities and expertise to provide high quality sequence and environmental (contextual) data for a broad range of marine environments (explored by e.g. Tara-Oceans, Malaspina, LTER sites, European marine stations)
b. Detailed knowledge on oceanographic databases, earth observation and monitoring as well as data management
c. Development of standards for describing sampling, sample handling and data processing in the context of molecular studies, also in marine environments
d. Expertise in the development of innovative bioinformatic approaches for data processing, analysis and integration, including developing professional software products
e. Expertise in ecosystems biology to allow interpretation of data and empower modelling approaches to enhance predictive capacities for the marine ecosystem
f. Expertise and state-of-the-art facilities to discover enzymatic functions and bioactive compounds for biotransformation and biocatalysis, including companies with emerging biotechnological products for white, green and red biotechnology
g. Expertise to develop IP agreements for pre-competitive microbial research materials and data as well as for the exploitation of high potential commercial applications provided by legal experts with a strong support by the Mediterranean Science Commission (CIESM) and EuropaBio
h. Exemplary training and outreach activities to support knowledge and technology transfer as well as capacity building around Europe. This will be supported by companies with a long standing expertise in providing workshops and training modules for European researchers as well as by the Ocean Sampling Day as a major event to measure and implement advances made in the project.
i. An experienced management and coordination team that will ensure professional management of the project in a collaborative working environment. It will implement performance control procedures to deliver in time and an appropriate strategy for the use of knowledge, dissemination and intellectual property rights management.
2. As targeted objectives, Micro B3 plans to deliver:
a. A comprehensive registry of European study sites and data sources for data safeguarding and acquisition
b. A set of community agreed standards for sampling and data acquisition (lab protocols), storage and exchange of data to reach a new level of interoperability and data integration across disciplines
c. Innovative software approaches for quality management, data processing, data integration, accessibility and visualisation
d. Ecosystem models for selected sites in the marine system to provide a predictive understanding of the contributions of functional microbial biodiversity to marine ecosystems functioning, with a special focus on the role of microbes to climate change and the effect of climate change on microbial communities.
e. A series of new biocatalytic processes, enzymes, biosynthetic pathway and bioactive compounds for use in biotechnological applications.
f. An innovative legal framework and model contracts for the protection and sustainable use of marine genetic resources.
g. A series of workshops, outreach and training entities as well as an Ocean Sampling Day to establish a baseline and make the project results accessible for researchers, the industry, the public and policy makers.
h. A new generation of marine microbial researchers well trained in oceanography, data management, omics technologies and biodiversity policy.
i. A new communication culture with bilateral understanding between computer scientists, biologists, biochemists, bioinformaticians, oceanographers as well as philosophy and judiciary.


